Motivation
Natural products are a major source of novel drugs, and with the rise of antibiotic resistance, there is an urgent need to discover new compounds. Genome mining enables the rapid identification of biosynthetic gene clusters (BGCs) responsible for natural product biosynthesis. Predicting the structures of the synthesized compounds is key to guiding their targeted discovery, but this requires detailed knowledge of the enzymatic reactions at each step of the biosynthetic pathway. While the core scaffold of many natural products can often be predicted from the initial biosynthetic steps, later tailoring modifications remain difficult to model due to the limited characterization of the enzymes involved. Reliable reaction prediction requires first gathering functional annotations and performing evolutionary analysis of these enzymes—only then can accurate computational predictions be made. In the absence of automated tools, this process becomes time-consuming. Although phylogenetic trees with functional annotations are occasionally published, reusing them directly is labor-intensive and technically challenging.
PhyloNaP addresses this gap by providing a centralized collection of annotated phylogenetic trees for enzymes involved in natural product biosynthesis.
Recent Updates
October 20, 2025
New Feature: Personal Tree Visualization
Users can now explore and analyze their own annotated phylogenetic trees directly in the PhyloNaP interface. This new functionality allows researchers to:
- Upload their own tree files (Newick format) along with metadata
- Visualize trees using the same interactive interface as the database entries
- Analyze clades using the “Get the summary of the clade” function
- If the annotation file includes a
MIBIGorMITEcolumn with valid IDs, corresponding molecular structures can also be displayed - Enjoy private browsing — uploaded trees and metadata are not stored on the server
Access this feature through the View page to start analyzing your phylogenetic data with PhyloNaP's advanced visualization tools.
1. Database Content Overview
What's in the Database
PhyloNaP's database contains comprehensive phylogenetic datasets for protein families involved in natural product biosynthesis. Each dataset includes:
- Phylogenetic tree with evolutionary model
- Sequence files in FASTA format
- Multiple sequence alignments
- Annotation files with functional data
- Optional elements: descriptions, superfamily classifications, reaction pathway images, and natural product structures
Database Composition
Curated Datasets (10)
High-quality, manually reviewed datasets from:
- Supplementary data or metadata from published articles
- Directly obtained from authors
- Provided by collaborators
Automated Datasets (~18,500)
Computationally generated phylogenetic trees covering:
- Broad enzyme family coverage
- Standardized quality control
- Consistent annotation pipeline
Database Generation Pipeline
Pipeline Steps
A) Data Collection
Protein sequences and annotations gathered from multiple high-quality databases:
- MiBIG: Characterized biosynthetic gene clusters
- MITE: Tailoring enzymes with experimentally proven reactions
- antiSMASH DB: Predicted biosynthetic gene clusters
- UniProt SwissProt: Curated protein annotations
Collected data includes taxonomic information, BGC types, functional annotations, and structural data (SMILES) when available.
B) Sequence Clustering
Sequences clustered using mmseqs easy-linclust with default
parameters (E-value: 1.000E-03) to group related proteins.
C) Quality Filtering
Clusters are filtered to remove:
- Explicitly eukaryotic proteins or SwissProt-only clusters
- Small clusters (<20 sequences)
- Sequences with unusual lengths (<150 or >1000 amino acids)
- NRPS/PKS sequences (require domain-level analysis)
D) Phylogenetic Analysis
- Alignment: Sequences aligned with
MAFFT auto - Trimming: Alignments trimmed with
TrimAl auto - Deduplication: Identical sequences removed (annotations preserved)
- Tree construction: Phylogenetic trees built with
FastTree - Rooting: Trees rooted using
MAD-root
E) Functional Classification
All sequences classified into superfamilies using HMMER with
superfamilies_1.75 profiles for consistent functional annotation.
Quality Assurance
Each step includes quality control measures to ensure reliable phylogenetic reconstructions and meaningful functional predictions.
Database Navigation
Users can efficiently explore the database using multiple filtering and sorting options:

- HMM name: Focus on specific enzyme families
- Source Distinguishes manually generated and annotated datasers and those generated by PhyloNaP pipeline
- Data type The automatic pipeline include only full proteins, but we kept an option for long multidomain proteins to load the datasets on specific domains
- N of sequences: The overall dataset size
- N of characterized: Number of proteins with known reaction (Proteins sourced from SwissProt+having Rhea number or from MITE)
- N of validated NP: Number of proteins from BGC, synthesizing characterized natural product (an entry from MiBiG)
- N of predicted NP: Number of proteins from predicted BGC (antiSMASHdb)
2. Examining Phylogenetic Tree Pages
Interactive Tree Visualization
PhyloNaP provides powerful interactive tools for exploring phylogenetic relationships and functional annotations. Each tree page combines evolutionary context with biochemical information to facilitate enzyme function prediction.
Explore the Tree

Summary of the clade feature
When you click on a node and select "Get the summary of the clade", a metadata summary is displayed. This summary shows all features associated with the selected node, and the descendant branches of that node will be highlighted in color.
By default, the features are sorted from those with the most identical values to those with the greatest diversity. However, users can adjust the sorting order manually using the arrow buttons. :

3. Enzyme Classification & Protein Placement
Protein Placement Pipeline Overview
PhyloNaP enables users to classify their protein sequences by placing them onto curated phylogenetic trees using a robust, multi-step computational pipeline.
Input Format
- Submit one or multiple protein sequences in FASTA format.
Similarity Search
- Each query is searched against a non-redundant PhyloNaP protein database using MMseqs2.
- The non-redundant set is built by aggregating all proteins and filtering out those with >70% similarity.
- Search thresholds:
- ≥ 30% sequence identity
- ≥ 50% alignment coverage
Placement Procedure
- For each query with hits, the associated dataset is retrieved.
- The query is aligned to the reference multiple sequence alignment used for the tree.
- The combined alignment is submitted to EPA-ng for placement onto the existing phylogenetic tree.
EPA-ng Placement Details
- EPA-ng uses the reference tree’s evolutionary model to generate a new tree from the extended alignment.
- It compares the new topology with the original to determine the most likely placement(s).
- Each placement includes:
- Placement position (clade or node)
- Branch lengths
- Likelihood weight ratio (LWR, 0–1; sum of all LWRs is 1)
- Note: A query may be placed in multiple locations, especially if it is distant from known clades or tree topology changes significantly, indicating higher uncertainty.
Understanding Branch Length & Placement Quality
Each placement features a pendant length—the branch connecting the query to the placement node/leaf.
- A long pendant length suggests greater evolutionary distance from the placement clade.
- This may indicate the query is not well represented by the clade’s features; interpret with caution.
Results Table

- Displays all matching datasets.
- Shows the Likelihood weight ratio of the best placement for each.
- If a query matches multiple trees, only the placement with the shortest pendant length is shown by default.
- You can toggle to display all alternative placements.
- Datasets are highlighted in red when the best placement has a pendant length > 1.
Tree Visualization
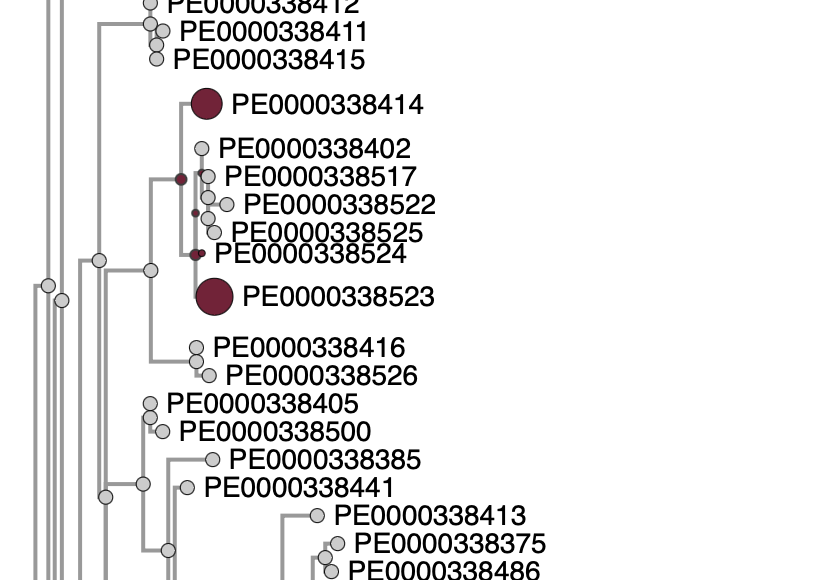
- Placements are shown as red circles on the phylogenetic tree.
- Circle size reflects the relative likelihood of each placement.
- If there is one dominant placement, only that is shown by default; others can be revealed by clicking “Show all placements”.
Contact and legal
Contact
Mail: For any questions or feedback, please contact us at aleksandra.korenskaia@uni-tuebingen.de
Ziemert Lab, University of Tübingen, Germany
Legal
Data Usage & Privacy
License:
PhyloNaP is released under the MIT License. The software is free to use for academic and commercial purposes.
Data Privacy:
- Submitted sequences are processed temporarily
- Job results are kept for 30 days
- No personal data is collected beyond optional email
Cookies:
This site uses minimal functional cookies to maintain session state. No tracking or analytics cookies are used.